An introduction to neuroinformatics

Meetup Details
The goal of brain imaging is to provide in-vivo measures of the human brain to better understand how the brain is structured, connected and functions. In this talk, we will discuss how to analyze brain imaging data in order to make sense of the large amount of data that comes out of the scanner.
This is the only session with the analysis all conducted with Python programming.
About the Speaker
Dr. Camille Maumet, is a research scientist in neuroinformatics at Inria, Univ Rennes, CNRS, Inserm in Rennes, France. Her research focuses on the variability of analytical pipelines and its impact on our ability to reuse brain imaging datasets. She obtained her PhD in computer science at the University of Rennes on the analyses of clinical neuroimaging datasets in functional magnetic resonance imaging and arterial spin labelling.
She was then a postdoctoral research fellow in the Institute of Digital Healthcare at the University of Warwick and the University of Oxford, where she focused on meta-analyses and standards for neuroimaging data sharing. She is also an open science advocate, involved in the development of more inclusive research practices and community-led research and participates in many collaborative efforts including Brainhack, the INCF, and the Open Science Special Interest Group of the Organization for Human Brain Mapping that she chaired in 2020.
Contact Speaker
An Introduction to Neuroinformatics
What is neuroinformatics
Neuroinformatics is a research field devoted to the development of neuroscience data and knowledge bases together with computational models and analytical tools for sharing, integration, and analysis of experimental data and advancement of theories about the nervous system function. In the INCF context, neuroinformatics refers to scientific information about primary experimental data, ontology, metadata, analytical tools, and computational models of the nervous system. The primary data includes experiments and experimental conditions concerning the genomic, molecular, structural, cellular, networks, systems and behavioural level, in all species and preparations in both the normal and disordered states.
Neuromatics Techniques
Make sure to convert the links to English if you are not French speaking
There are many techniques Camille uses for brain imaging. The techniques she uses are Magnetic Resonance imaging (MRI), Functional near-infrared spectroscopy (fNIRS), Electoencephalography (EEG). However, she mostly uses MRI which generates certain data from brain imaging which gives information about the brain such as anatomy, connectivity, function, brain mapping.
Anatomy
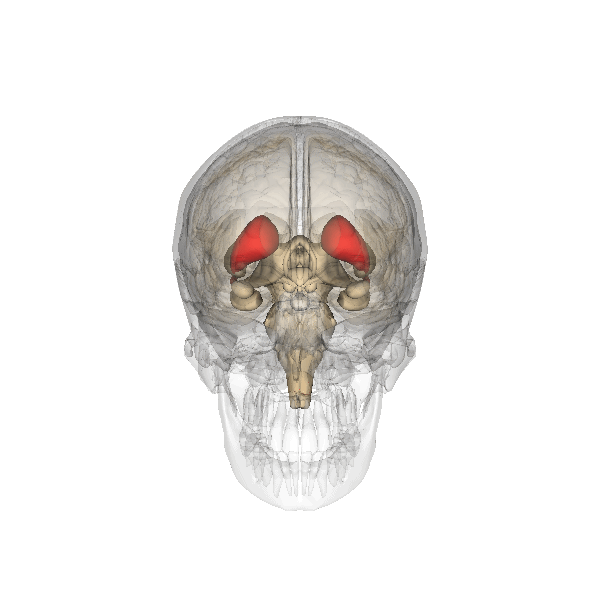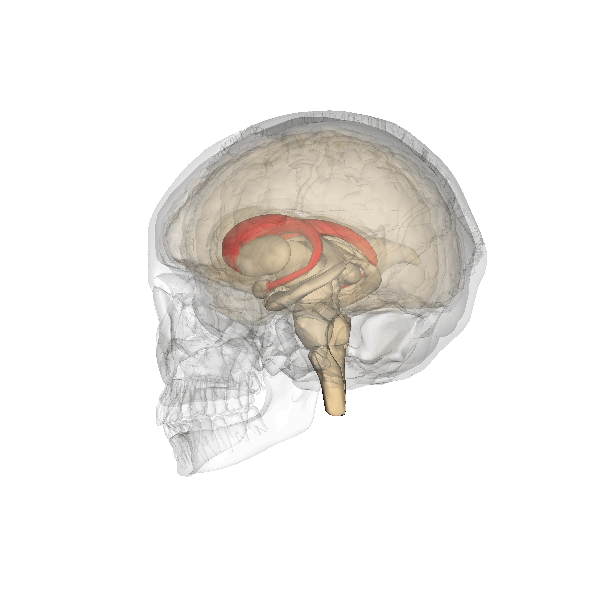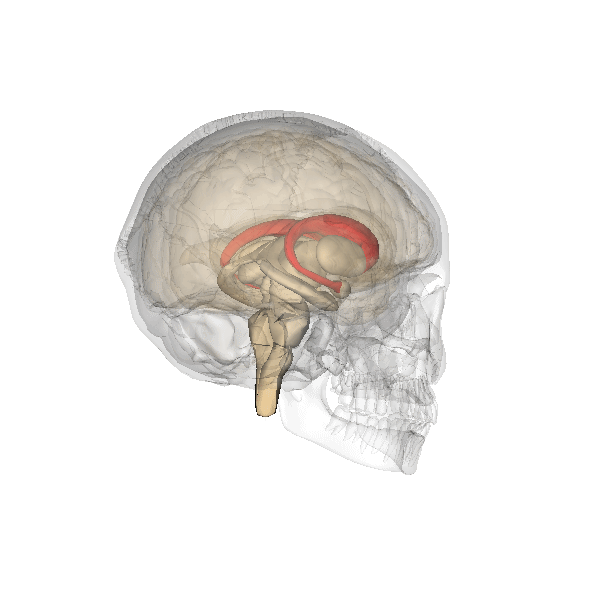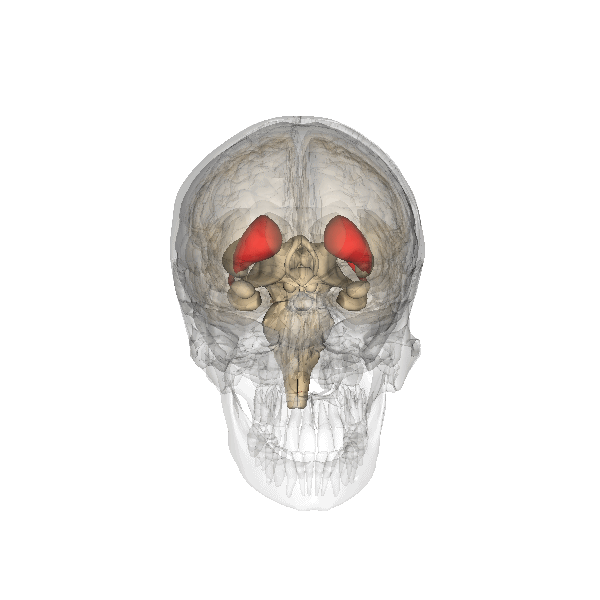
Camille explains looking inside the Brain is like looking into the earths core. Song et al 2021 Song et al (2021) shows images of the Cortical Surfaces (figure below) and details its different areas.
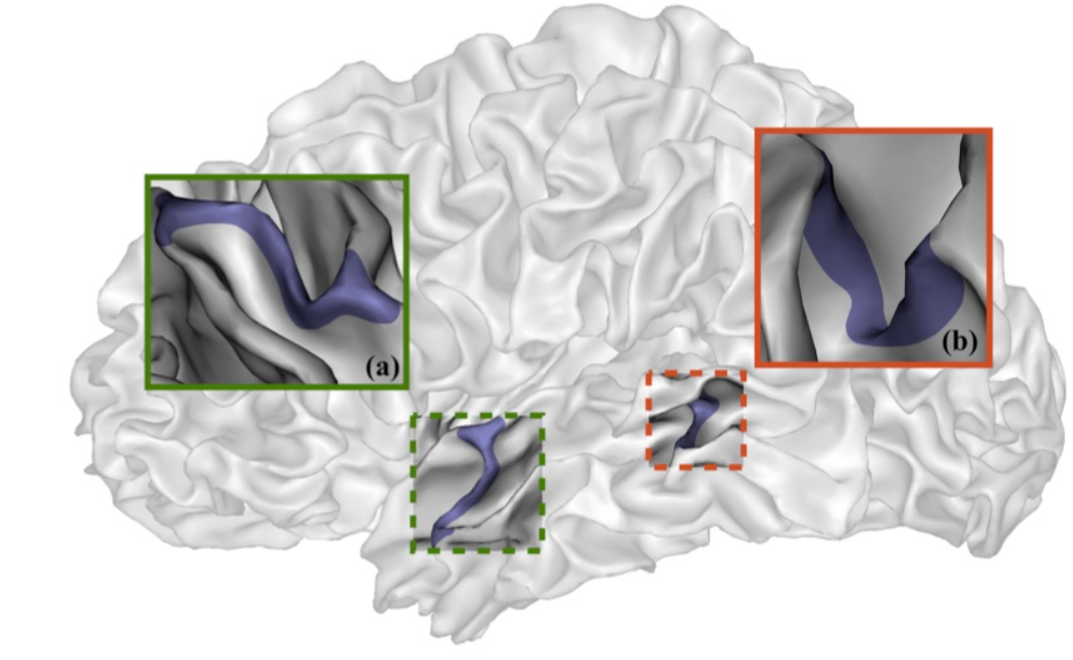
Connectivity
Another information to look at is the structural Connectivity within the brain. The connectivity in the Brain can be compared as a street map. The road map allows us to see the pathways within the Brain.
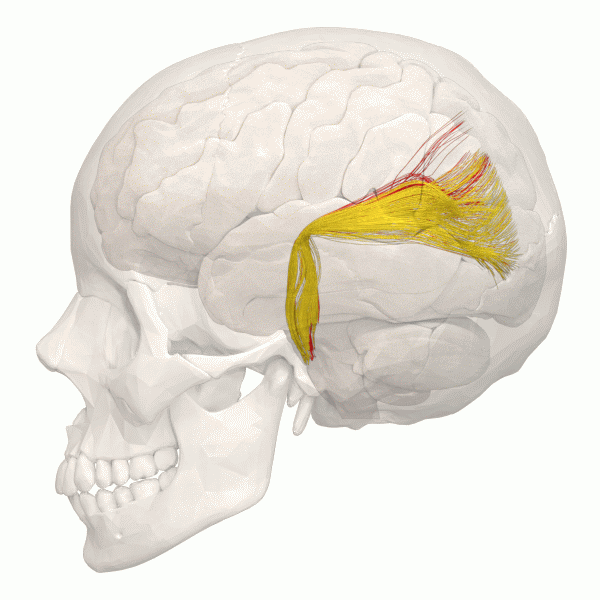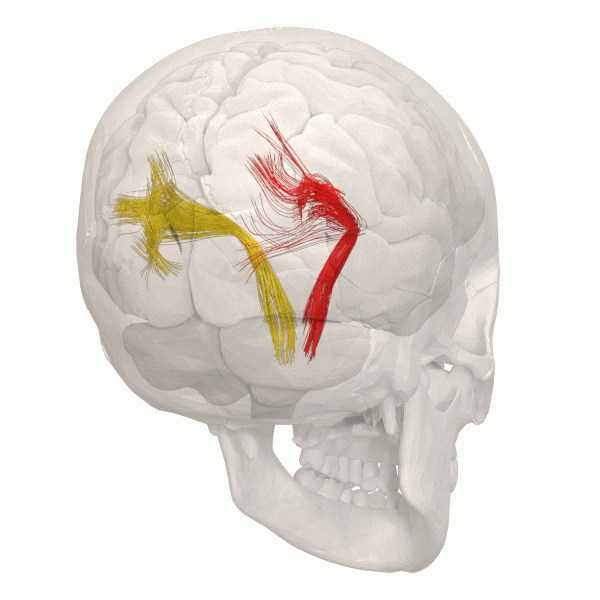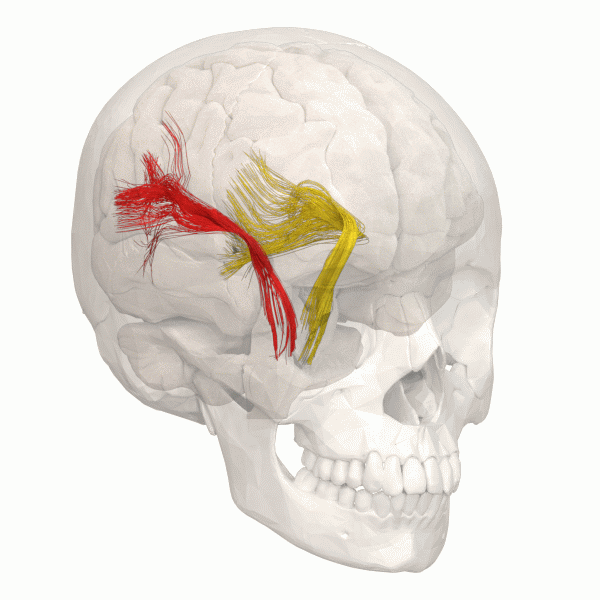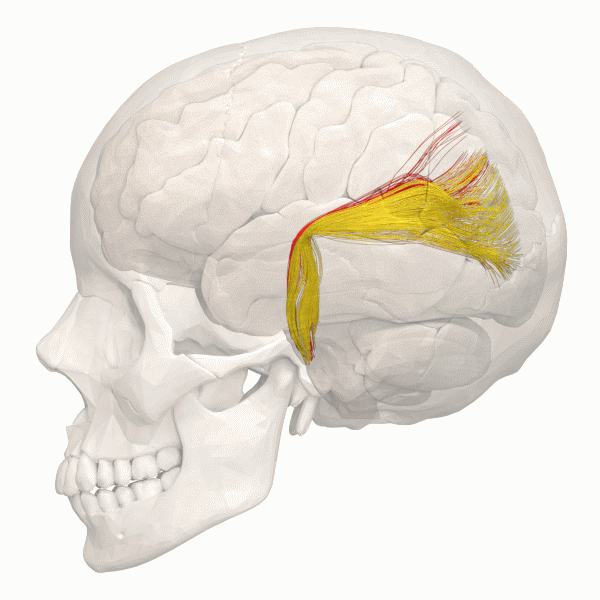
The information gained can be simulated using using Python.
1. Demonstration of : exploring fMRI data
Exploring fMRI can be done using;
Nibabel to Access a cacophony of neuro-imaging file formats
Nilearn enables approachable and versatile analyses of brain volumes. It provides statistical and machine-learning tools, with instructive documentation & open community.
It supports general linear model (GLM) based analysis and leverages the scikit-learn Python toolbox for multivariate statistics with applications such as predictive modelling, classification, decoding, or connectivity analysis.
repronim/neurodocker Neurodocker is a command-line program that generates custom Dockerfiles and Singularity recipes for neuroimaging and minifies existing containers. See the website https://www.repronim.org/neurodocker for more information.
2. Demonstration on Motion Correction
Exploring Motion Correction can be done using.
Statistical Parametric Mapping refers to the construction and assessment of spatially extended statistical processes used to test hypotheses about functional imaging data. These ideas have been instantiated in a free and open source software that is called SPM. The SPM software package has been designed for the analysis of brain imaging data sequences. The sequences can be a series of images from different cohorts, or time-series from the same subject. The current release is designed for the analysis of fMRI, PET, SPECT, EEG and MEG.
Nipype, an open-source, community-developed initiative under the umbrella of NiPy, is a Python project that provides a uniform interface to existing neuroimaging software and facilitates interaction between these packages within a single workflow.
Nipype provides an environment that encourages interactive exploration of algorithms from different packages (e.g., ANTS, SPM, FSL, FreeSurfer, Camino, MRtrix, MNE, AFNI, Slicer, DIPY), eases the design of workflows within and between packages, and reduces the learning curve necessary to use different packages. Nipype is creating a collaborative platform for neuroimaging software development in a high-level language and addressing limitations of existing pipeline systems.
Nipype allows you to:
easily interact with tools from different software packages
combine processing steps from different software packages
develop new workflows faster by reusing common steps from old ones
process data faster by running it in parallel on many cores/machines
make your research easily reproducible
share your processing workflows with the community
3. Demonstration on : Preprocessing
Reproduction of SPM tutorial using Nipype: https://gitlab.inria.fr/egermani/reproduced_tutorial. Statistical Parametric Mapping as well as Nipypr are also used for preprocessing demonstration.
You find slides and images following the GitHub link An Introduction to Neuromatics GitHub
With over 2,500 views on YouTube, this meetup session is the most popular R-Ladies Gaborone session with active participation during the meetup. The figure shows the comments on YouTube.
Resources
[Nipype Tutorial](https://miykael.github.io/nipype_tuto...) -- Annual Brain Imaging Events:
[OHBM Brainhack (Brain Hackathon) June 16-18](https://ohbm.github.io/hackathon2022/)[registration via](https://www.humanbrainmapping.org/i4a... )
[OHBM Open Science Room (Discussions around open science practices & brain imaging)](https://ohbm.github.io/osr2022/)
[Neuromatch Academic (Online course in computational neuroscience and deep learning)](https://neuromatch.io/)
Youtube Link
Watch the recording of the meetup session.